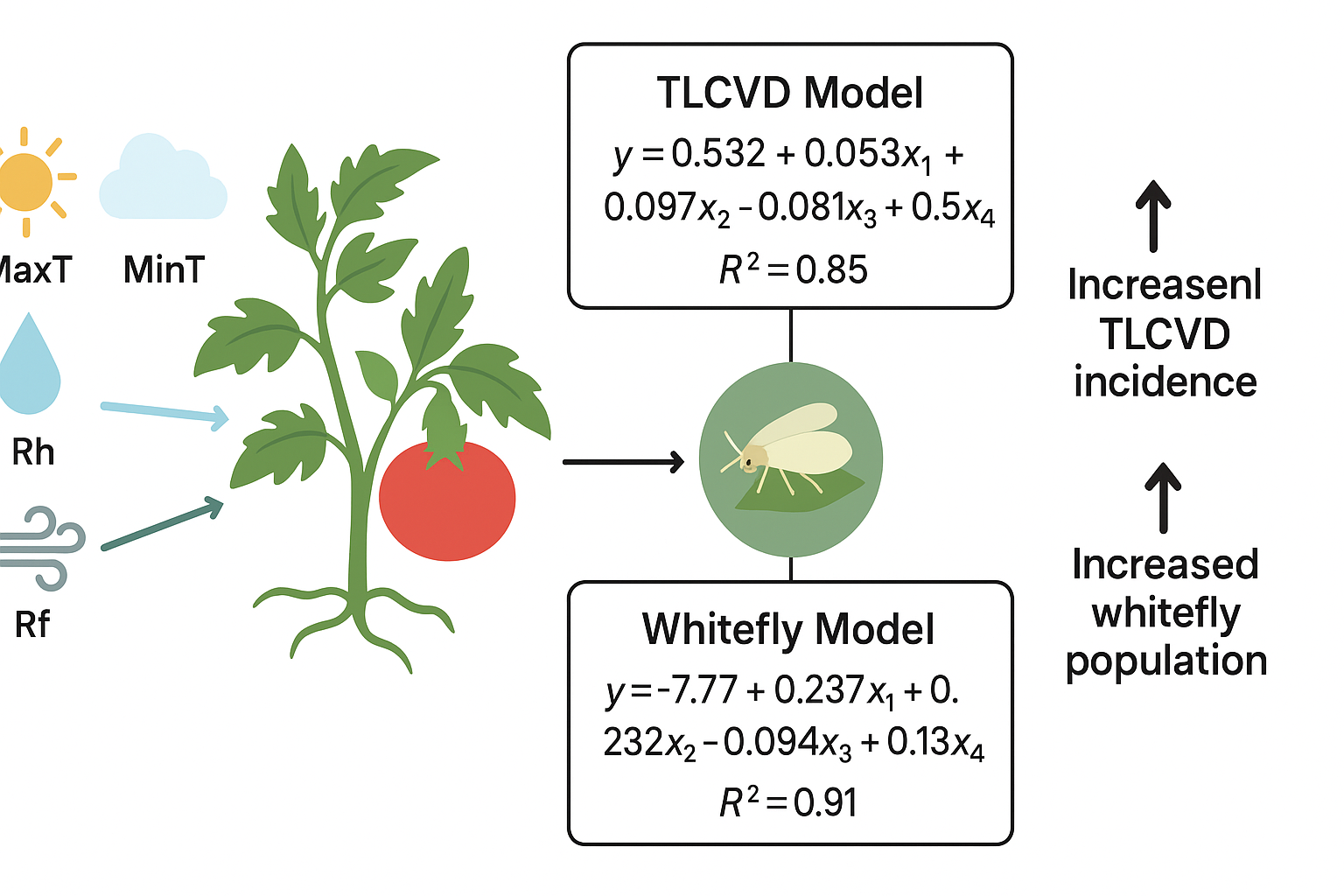
Models developed to predict diseases associated with the tomato leaf curl virus (TLCVD) in different countries of the world. The research has been set to identify those parameters that are maximally influenced by the infectious weather variables. Therefore, the objectives of this research were to forecast the TLCVD disease together with its vector whitefly based on meteorological conditions and to validate the two models. Sown in three replications using RCBD over two seasons were the following varieties of tomato genotypes: Big Beef, Sitara-TS-101, 014276, Caldera, and Salma. Data on disease incidence and weather variables were used to develop a disease-predictive model for TLCVD by employing stepwise regression analysis. The criterion of evaluation for this model was a comparison between predicted and empirical observations. TLCVD predictive model y= 0.532+ 0.053x1 (°C) +0.97x2 (°C) -0.081x3 (%) +0.15x4 (mm) R2= 0.85 y= TLCV disease, while whitefly predictive model y= -7.77+0.237x1 (°C) +0.223x2 (°C) -0.094x3 (%) +0.13x4 (°C) +0.085x5 (m/s) R2= 0.91. All weather parameters affected the incidence of the disease in addition to the infestation with the whitefly. A revised RMSE of 0.51 after exclusion of outliers demonstrated the accuracy in a determined range among different cultivars. Based on the current study findings, predictive models for TLCV disease and in respect of whitefly would be developed for different agroecological zones of Pakistan.